Phytophthora insulinativitatica
|
Phytophthora spp. in subclade 2a: portion of the seven-loci ML phylogeny featuring the type cultures of 212 described species (by T. Bourret). Notice the position of P. insulinativitatica Ex-type CBS 146553. Gloria Abad, USDA S&T.
|
|
Phytophthora spp. in subclade 2a: Morphological Tabular key (PDF) and Tabular key legends (PDF) in IDphy2 KEY SECTION. Notice the data of P. insulinativitatica Ex-type CBS 146553. Gloria Abad, USDA S&T.
|
|
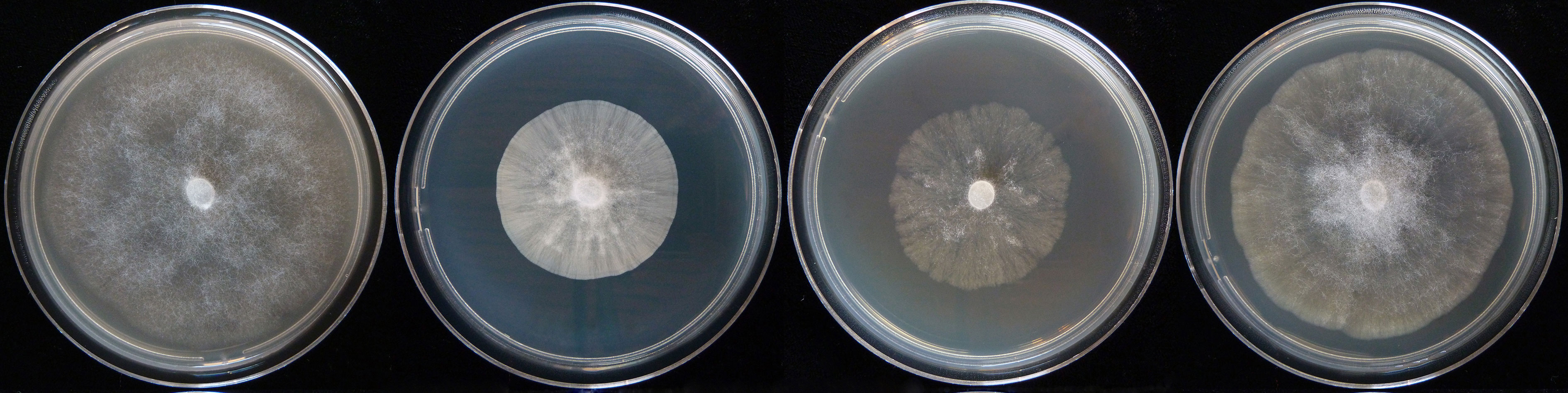
colony morphology after 7 days growth at 20°C on carrot agar, potato-dextrose agar, malt extract agar, and V8 agar (from left to right) |
|
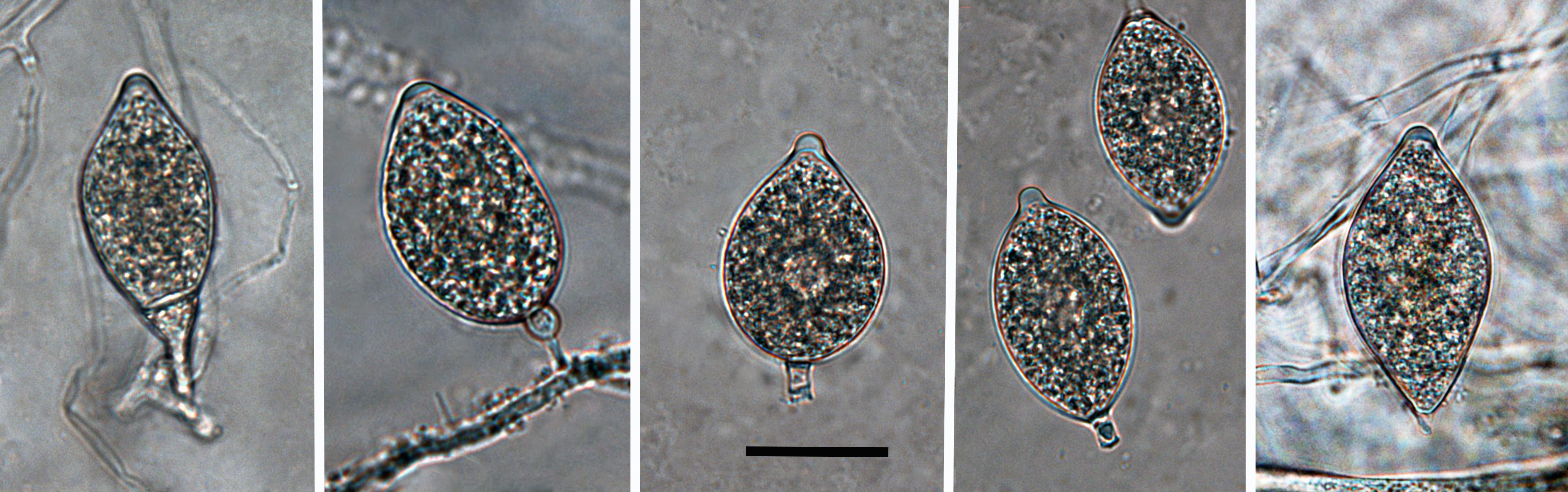
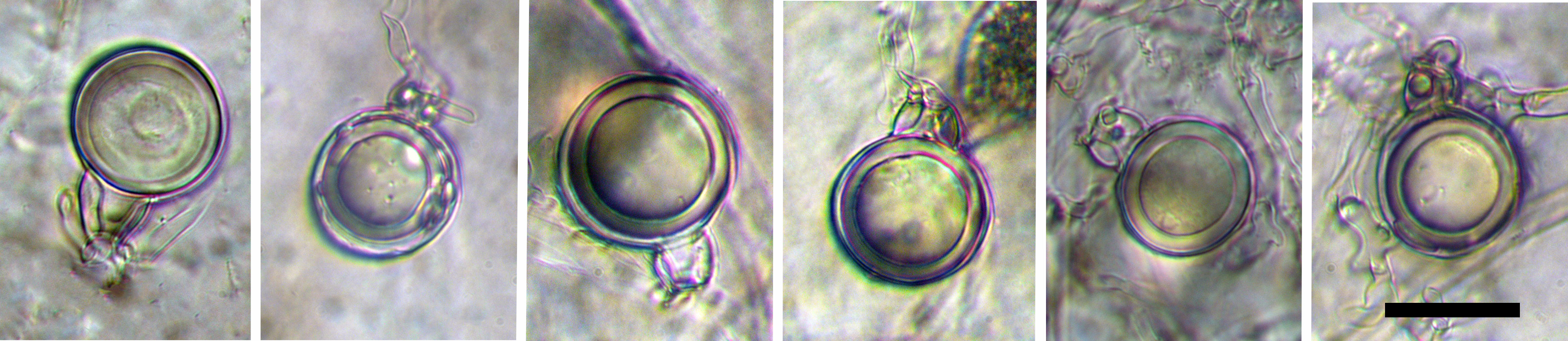
mostly caducous, limoniform, papillate to semi-papillate sporangia; scale bar = 25µm |
|
compound sympodia; scale bar = 40µm |
|
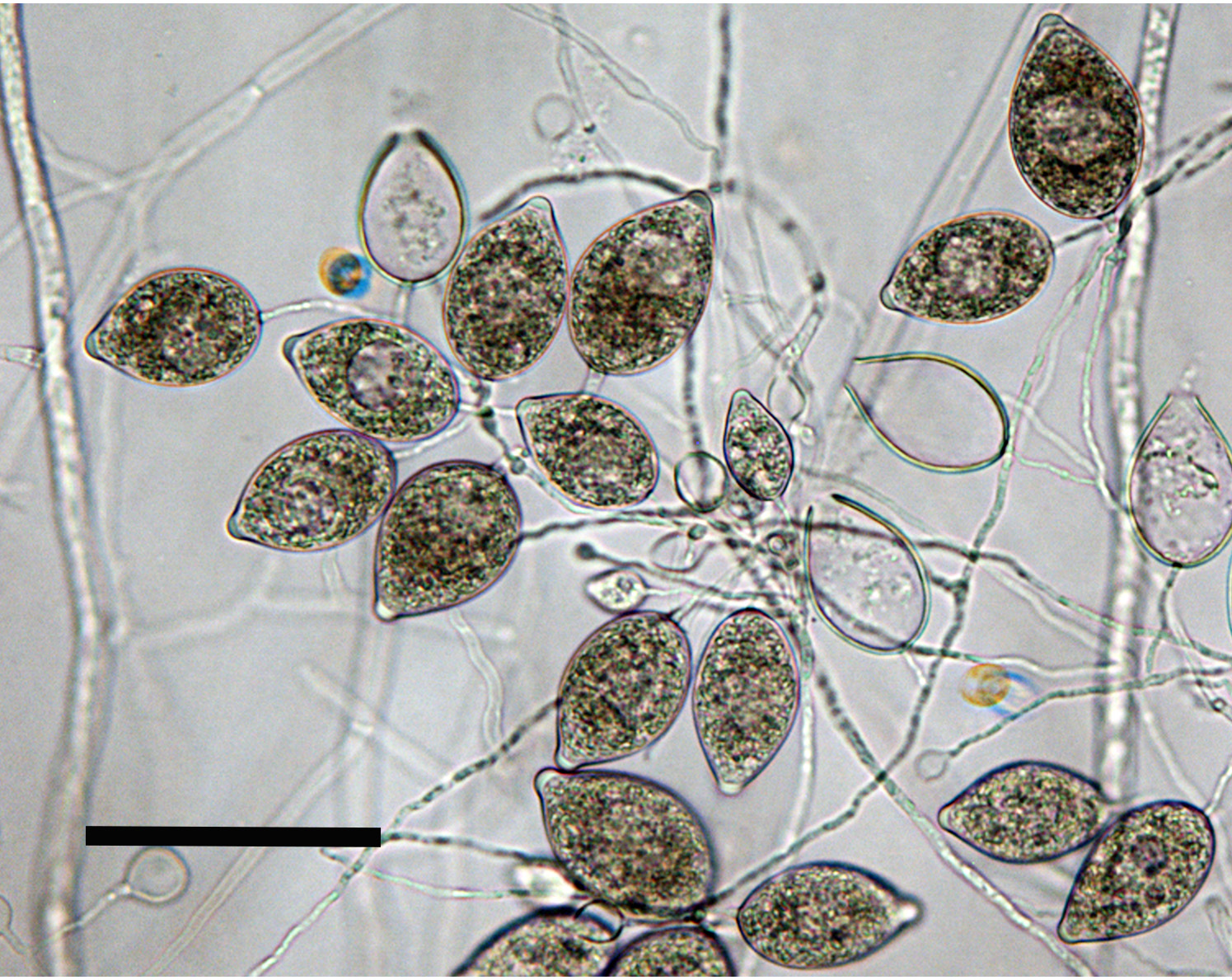
oogonia with smooth walls containing plerotic oospores; antheridia both amphigynous and paragynous; scale bar = 25µm |
Name and publication
Phytophthora insulinativitatica Q.N. Dang, G. Hardy and T.I. Burgess (2021)
Dang QN, Pham TQ, Arentz F, Hardy GE, Burgess TI. 2021. New Phytophthora species in cladeclade:
a taxonomic group of organisms classified together on the basis of homologous features traced to a common ancestor
2a from the Asia-Pacific region including a re-examination of P. colocasiae and P. meadii. Mycological Progress 20 (2): 111–129.
Corresponding author: T.Burgess@murdoch.edu.au
Nomenclature
Mycobank
Etymology
referring to the locality (Christmas Island) where this species was recovered
Typification
Type: AUSTRALIA, Christmas Island, rhizosphere soil from disturbed rainforest, March 2016, G. Hardy, holotype MURU468
Ex-type: culture CBS 146553
Sequences of ex-type in manuscript: ITS KY212028, β-tubulin MT583631, hsp90 MT583676, cox1 MT583646, cox2 MT583661, nadh1 MT583691, rps10 MT583707
Ex-type in other collections
(ET) CBS 146553, CI-10A, AUS2D
Molecular identification
Voucher sequences for barcoding genes (ITS rDNA and COI) of the ex-type (see Molecular protocols page)
Phytophthora insulinativitatica ITS rDNA, COI
Voucher sequences for Molecular Toolbox with seven genes (ITS, β-tub, COI, EF1α, HSP90, L10, and YPT1
(see Molecular protocols page) (In Progress)
Voucher sequences for Metabarcoding High-throughput Sequencing (HTS) Technologies [Molecular Operational Taxonomic Unit (MOTU)]
(see Molecular protocols page) (In Progress)
Sequences with multiple genes for ex-type in other sources
- NCBI: Phytophthora insulinativitatica
- EPPO-Q-bank: Phytophthora insulinativitatica
- BOLDSYSTEMS: Phytophthora insulinativitatica
Position in multigenic phylogeny with 7 genes (ITS, β-tub, COI, EF1α, HSP90, L10, and YPT1)
Clade clade:
a taxonomic group of organisms classified together on the basis of homologous features traced to a common ancestor
2a
Morphological identification
Colonies and cardinal temperatures
Colony colony:
assemblage of hyphae which usually develops form a single source and grows in a coordinated way
morphology smooth and uniform on PDA, cottony and uniform on CA, stellate on MEA, produced a pattern of concentric circles on V8A. Minimum growth temperature 4°C, optimum 30°C, and maximum 35°C.
Conditions for growth and sporulation
SporangiaSporangia:
sac within which zoospores form, especially when water is cooled to about 10°C below ambient temperature; in solid substrates, sporangia usually germinate by germ tubes
are produced in water cultures (soil extract) and not observed in solid media. OogoniaOogonia:
the female gametangium in which the oospore forms after fertilization by the antheridium
are formed readily in paired cultures on V8A after about 21 d.
Asexual phase
SporangiaSporangia:
sac within which zoospores form, especially when water is cooled to about 10°C below ambient temperature; in solid substrates, sporangia usually germinate by germ tubes
were semi-papillate, caducouscaducous:
pertaining to sporangia that become dislodged readily (i.e. deciduous) and separate from the sporangiophore (cf. persistent)
, and predominantly limoniform with no proliferationproliferation:
formation of a sporangium within an empty sporangium that has previously emitted zoospores (internal or nested) or after the sporangiophore has emerged from the empty sporangium (external)
. SporangiaSporangia:
sac within which zoospores form, especially when water is cooled to about 10°C below ambient temperature; in solid substrates, sporangia usually germinate by germ tubes
averaged 34.1 x 21.5 µm (overall range 25.4–45 x 16.1–28.8 µm). Sporangiophores umbellate and compound sympodiasympodia:
a type of sporangiophore which appears simple, but where each successive sporangium develops on a branch behind and to one side of the previous apex, where growth has already ceased
. Hyphal swellings present. ChlamydosporesChlamydospores:
an asexual spore with a thickened inner wall that is delimited from the mycelium by a septum; may be terminal or intercalary, and survives for long periods in soil
present.
Sexual phase
Heterothallic. OogoniaOogonia:
the female gametangium in which the oospore forms after fertilization by the antheridium
had smooth walls and size ranged from 20.3–32.5 µm. OosporesOospores:
zygote or thick-walled spore that forms within the oogonium after fertilization by the antheridium; may be long-lived
pleroticplerotic:
pertaining to an oospore that fills the oogonium (cf. aplerotic)
to semi-aplerotic, globoseglobose:
having a rounded form resembling that of a sphere
, size ranged 18.3–30.5 µm. AntheridiaAntheridia:
the male gametangium; a multinucleate, swollen hyphal tip affixed firmly to the wall of the female gametangium (the oogonium)
amphigynousamphigynous:
pertaining to the sexual stage in which the antheridium completely surrounds the stalk of the oogonium (cf. paragynous)
.
Most typical characters
Phytophthora insulinativitatica resides in cladeclade:
a taxonomic group of organisms classified together on the basis of homologous features traced to a common ancestor
2a most closely related to P. botryosa. The feature that most distinguishes P. insulinativitatica is the umbellate and compound symposia.
Additional specimen(s) evaluated
Australia, Christmas Island, rhizosphere soil from disturbed rainforest, 2016, G. Hardy, CBS 146553; CI21A
Hosts and distribution
Distribution: Australia (Christmas Island)
Substrate: rhizosphere soil
Disease note: no known disease
Host: disturbed rainforest vegetation
Additional references and links
- SMML USDA-ARS: Phytophthora insulinativitatica
- EPPO Global Database: Phytophthora insulinativitatica
- Forest Phytophthoras of the World: Phytophthora insulinativitatica
- CABI Digital Library: Phytophthora insulinativitatica
- Encyclopedia of Life (EOL): Phytophthora insulinativitatica
- Index Fungorum (IF): Phytophthora insulinativitatica
Fact sheet author
Treena Burgess, Ph.D., Phytophthora Science and Management, Harry Butler Institute, Murdoch University, Australia
Z. Gloria Abad, Ph.D., USDA-APHIS-PPQ-S&T Plant Pathogen Confirmatory Diagnostics Laboratory (PPCDL), United States of America.