Molecular identification
Molecular protocols
In order to facilitate the correct identification of Phytophthora species, we provide standards of procedures (SOPs) for all steps involved with molecular identification using sequencing analysis of the ITS rDNA and COI barcoding regions. Each SOP is available as a downloadable PDF.
SOP-PID-01.01 Molecular: Equipment, materials, and reagents for DNA extraction and PCR
SOP-PID-02.01 Molecular: DNA extraction for Phytophthora species using Qiagen kit (suggested)
SOP-PID-03.01 Molecular: Primer dilutions for PCR amplifications
SOP-PID-04.01 Molecular: Gels: electrophoresis, and photos
SOP-PID-05.05 Molecular: Identification of Phytophthora species with ITS rDNA and COI barcoding genes
All sequences submitted to NCBI by the Beltsville Lab for seven genes from the ex-types and other well-authenticated specimens can be found here. The seven genes are: internal transcribed spacer (ITS rDNA), elongation factor 1-alpha (EF1a), shock protein 90 (HSP90), beta-tubulin, 60S ribosomal protein L10 (RPL10), Ras-related protein ypt1 (ypt), and cytochrome c oxidase subunit 1 (COI), mitochondrial.
Sequences for new species not yet included in IDphy, as well as sequences for putative new species in progress, can be found here.
Identifying Phytophthora unknowns
For quick reference, here are the steps from SOP-PID-05 for sequence analysis of your unknown. Once you have your FASTA sequence, follow these steps to identify Phytophthora unknown species using vouchers of the ITS rDNA gene of 139 ex-types and 22 well-authenticated species. If this is your first time conducting this analysis, use the example sequences in SOP-PID-05 (page 6) to practice the steps for better understanding. The example below uses example sequence number 1.
-
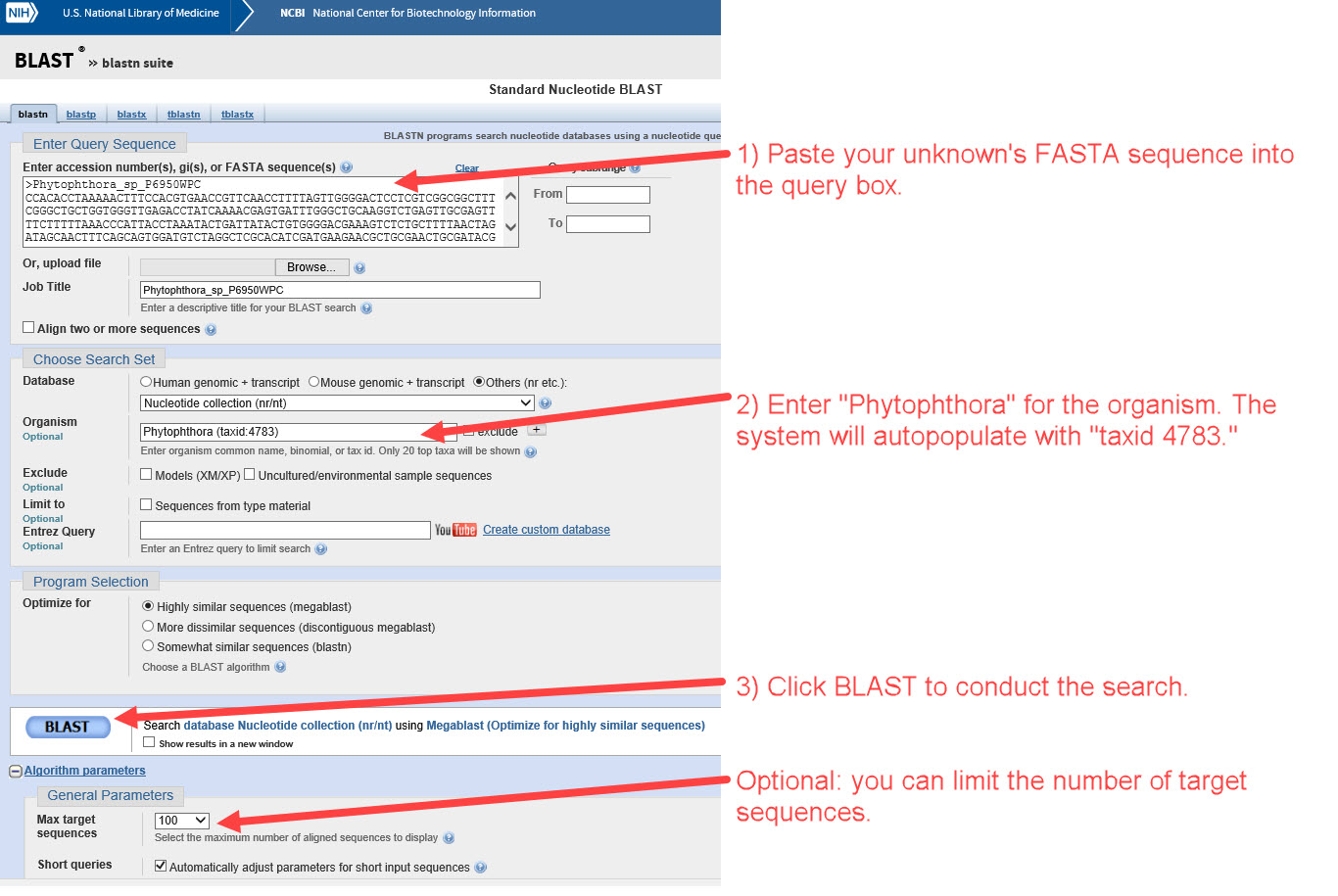
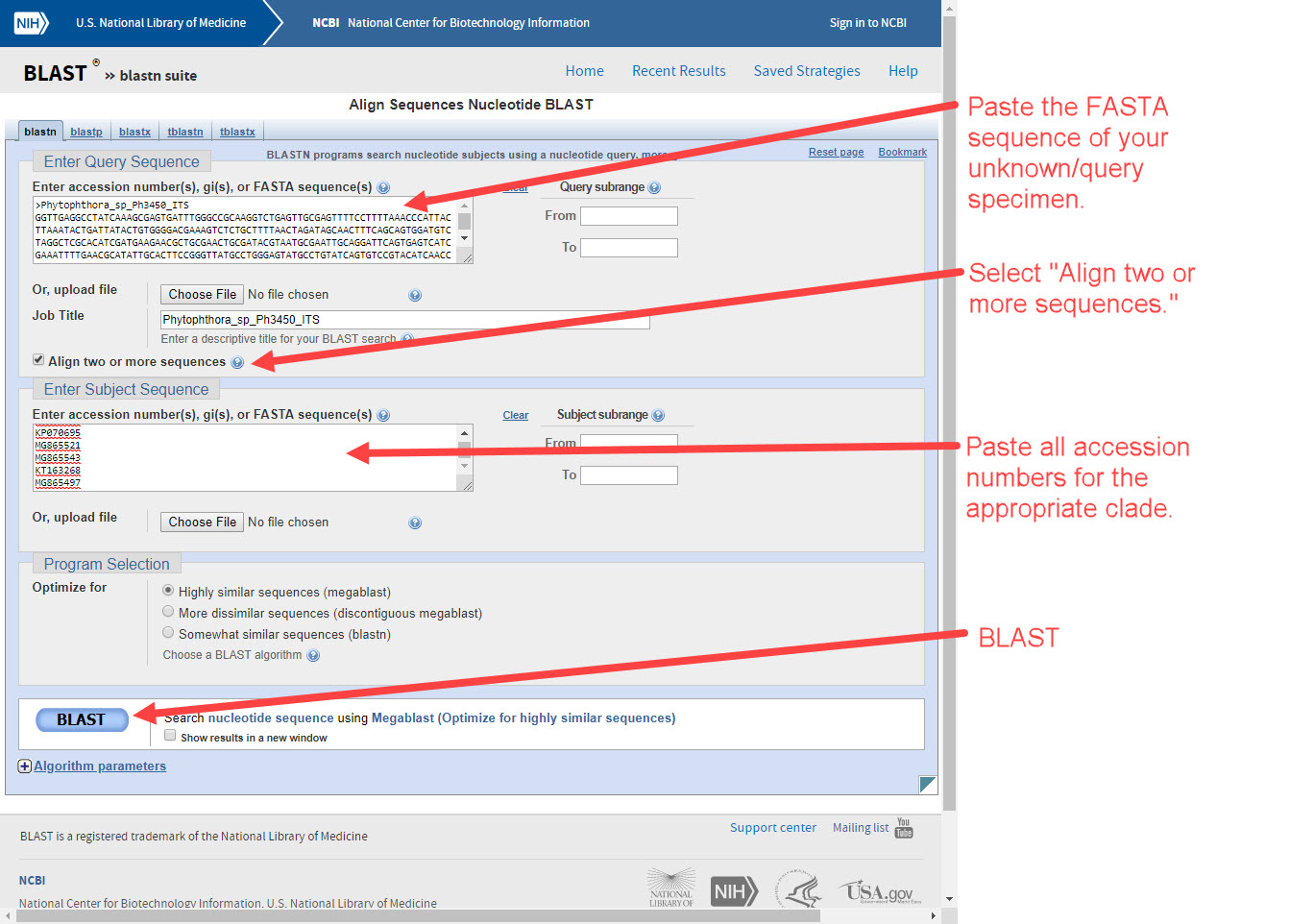
1) Go to NCBI BLAST blastn suite and perform a nucleotide BLAST analysis of the Phytophthora unknown's FASTA sequence for ITS rDNA or COI. -
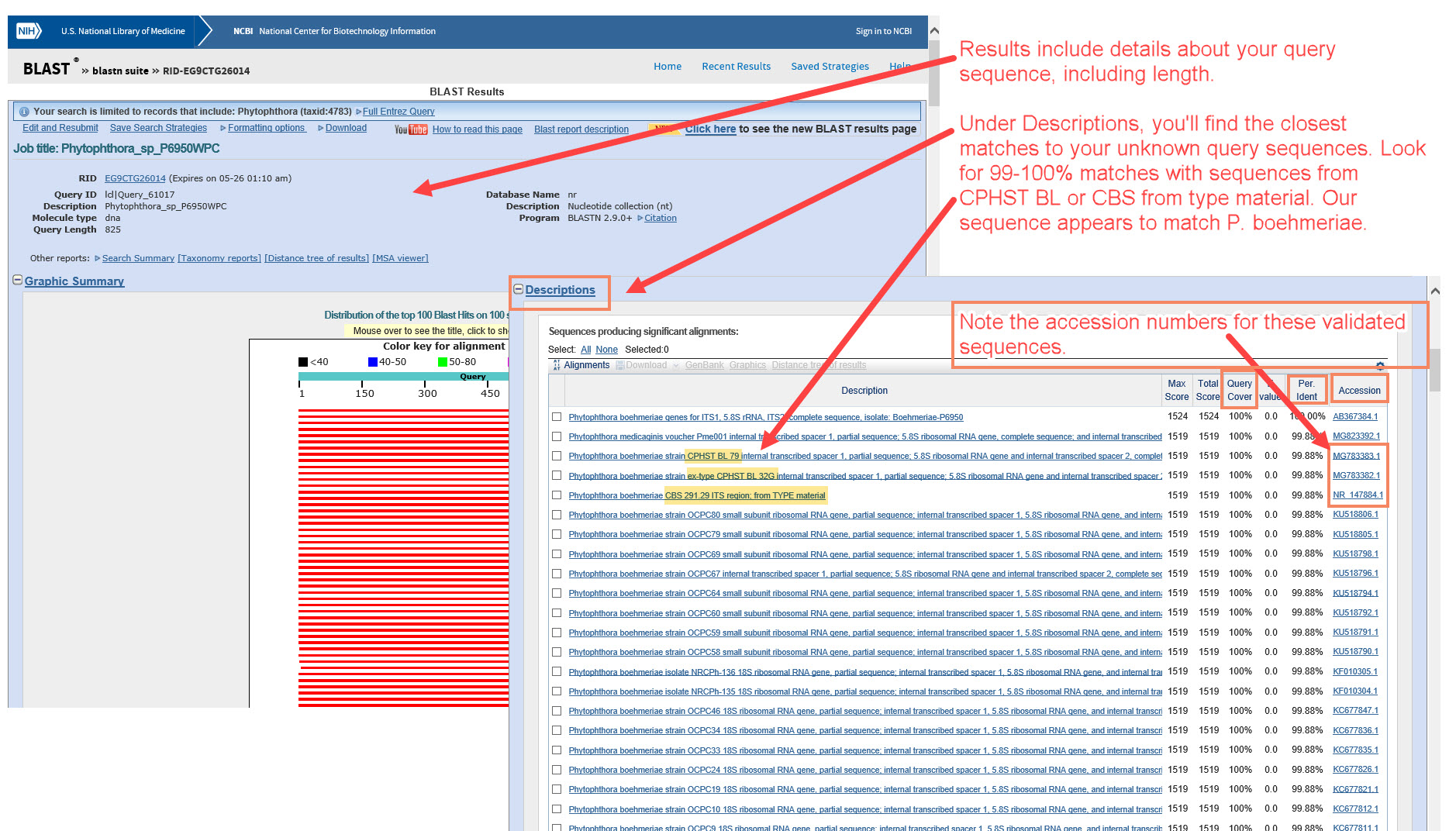
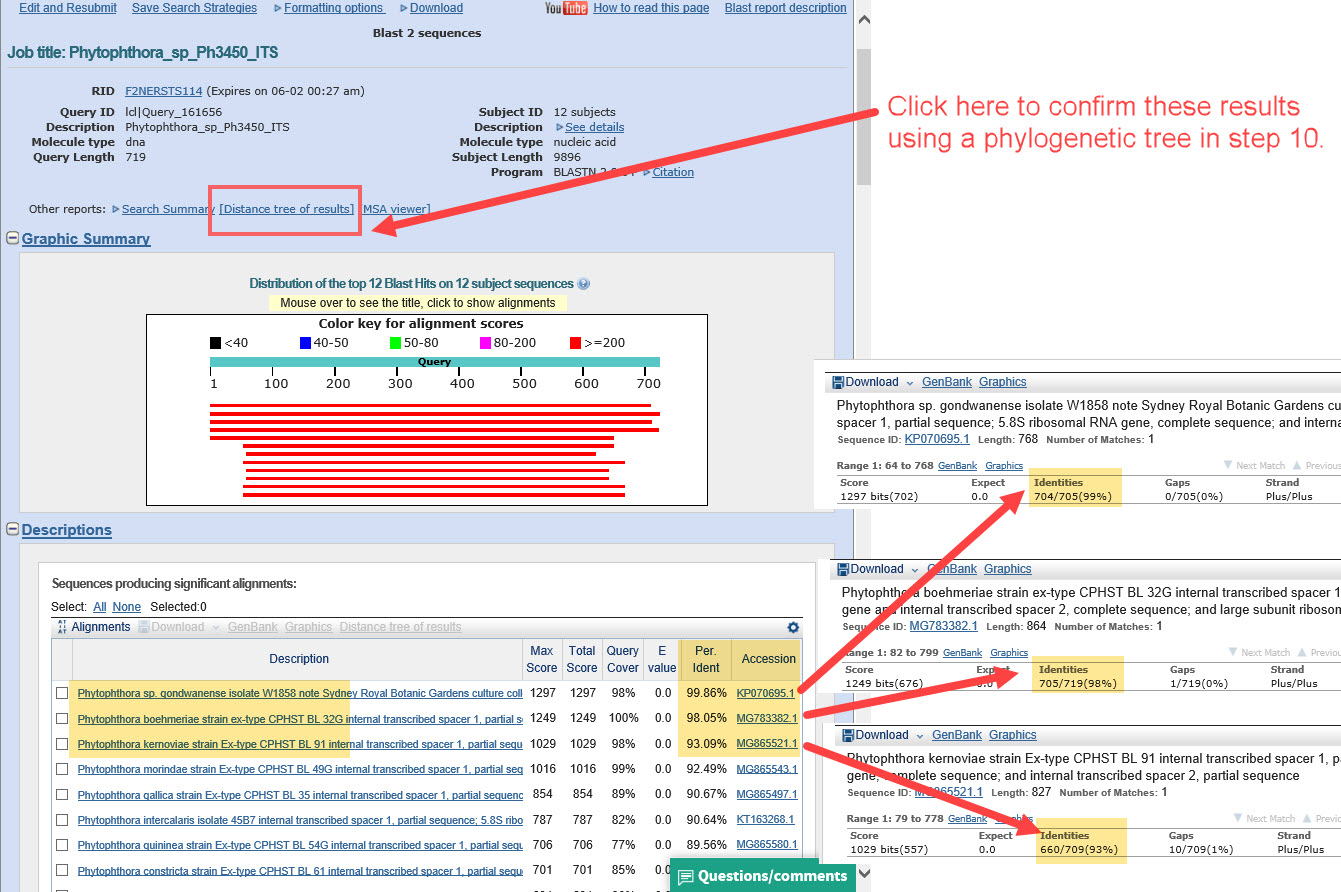
2) Under “Description”, look for the species that have the closest alignment(s) with your query/unknown sequence. This example appears to match P. boehmeriae. The next steps will help confirm whether the matching sequence is from a well-authenticated specimen. -
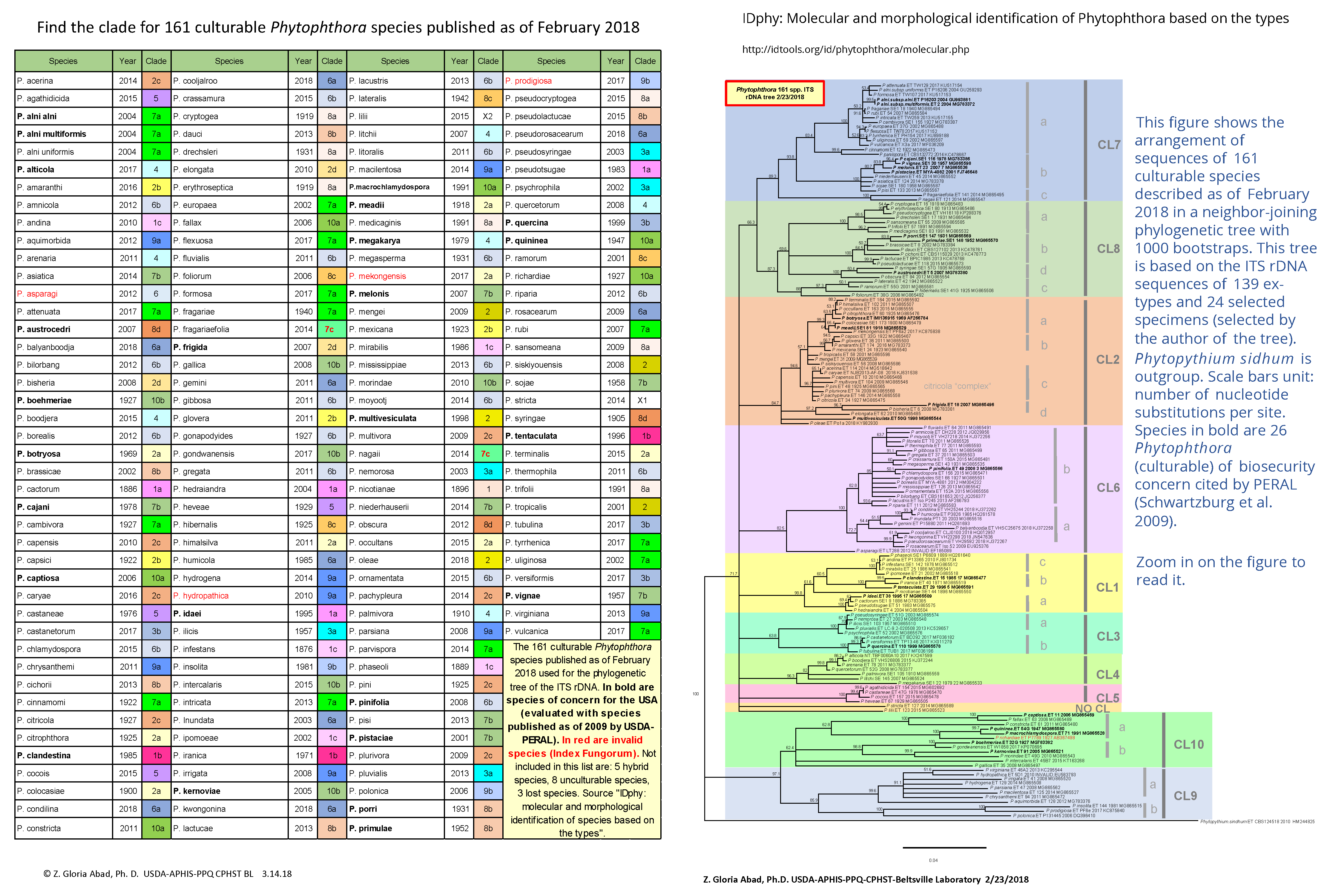
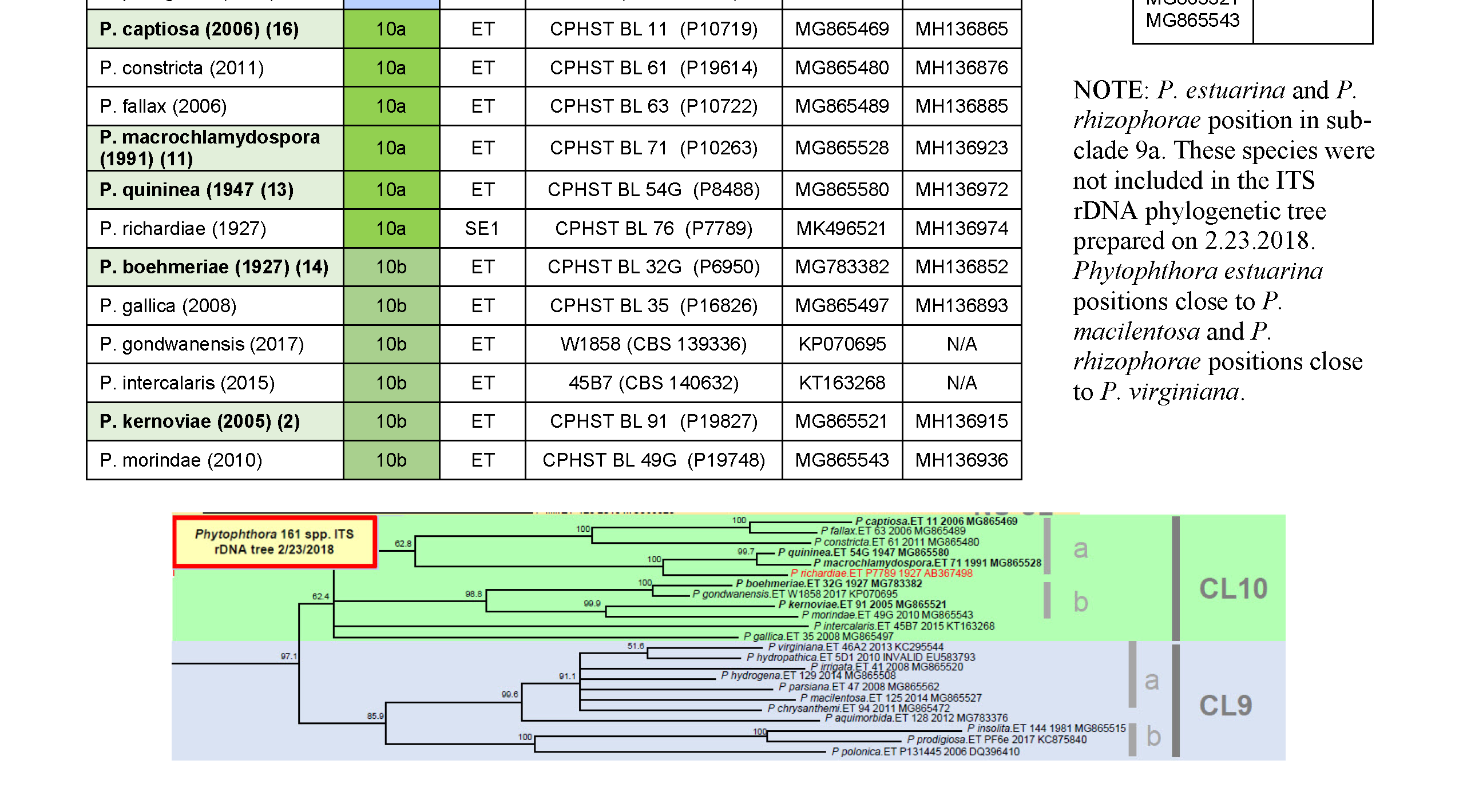
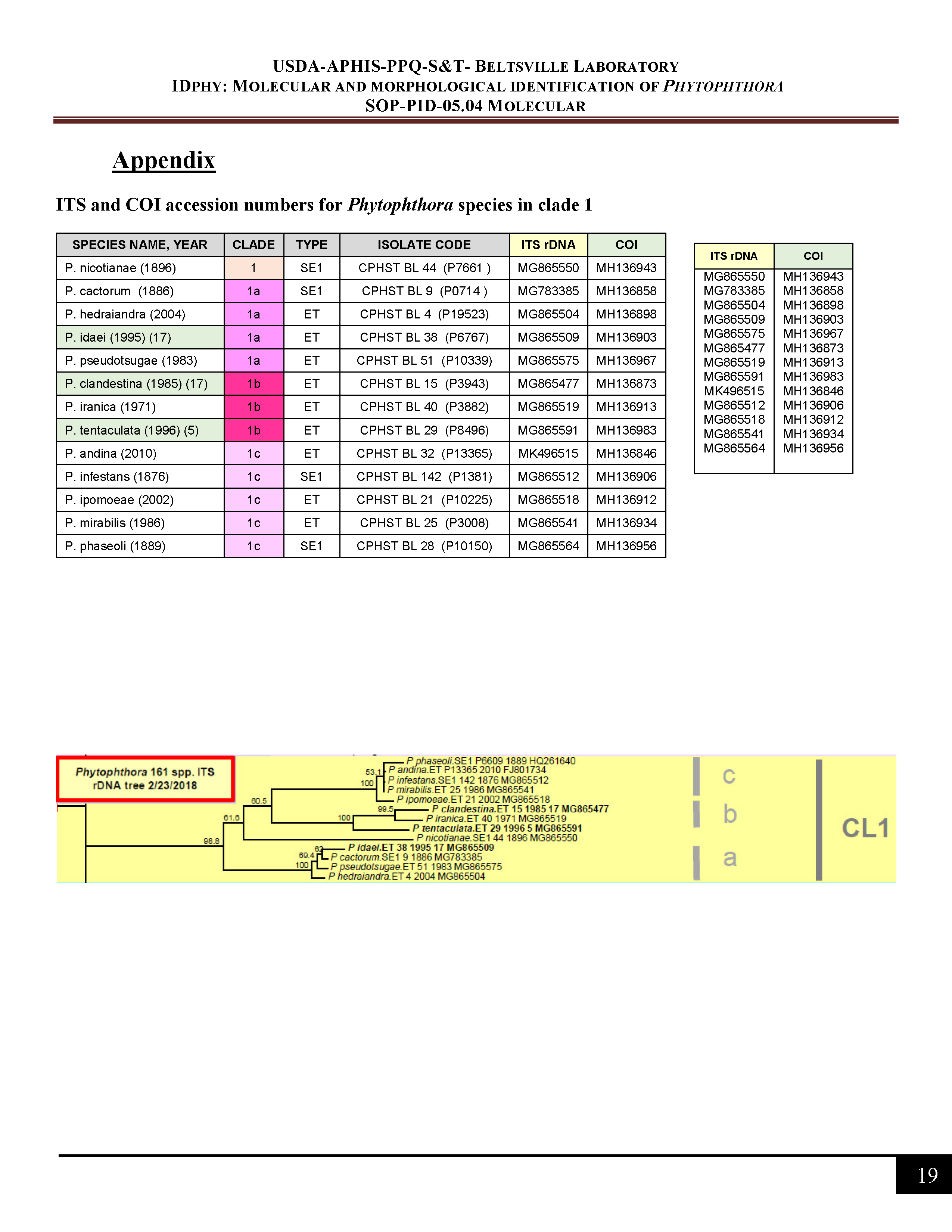
3) First, use the clade table to find the clade where the possible species positions. For this example, P. boehmeriae positions in clade 10b.
-
4) Once you have found the clade, check the appropriate accession number table (scroll down or check in the appendix of SOP-PID-05 for the tables) to find the accession number for the ex-type of the possible species. -
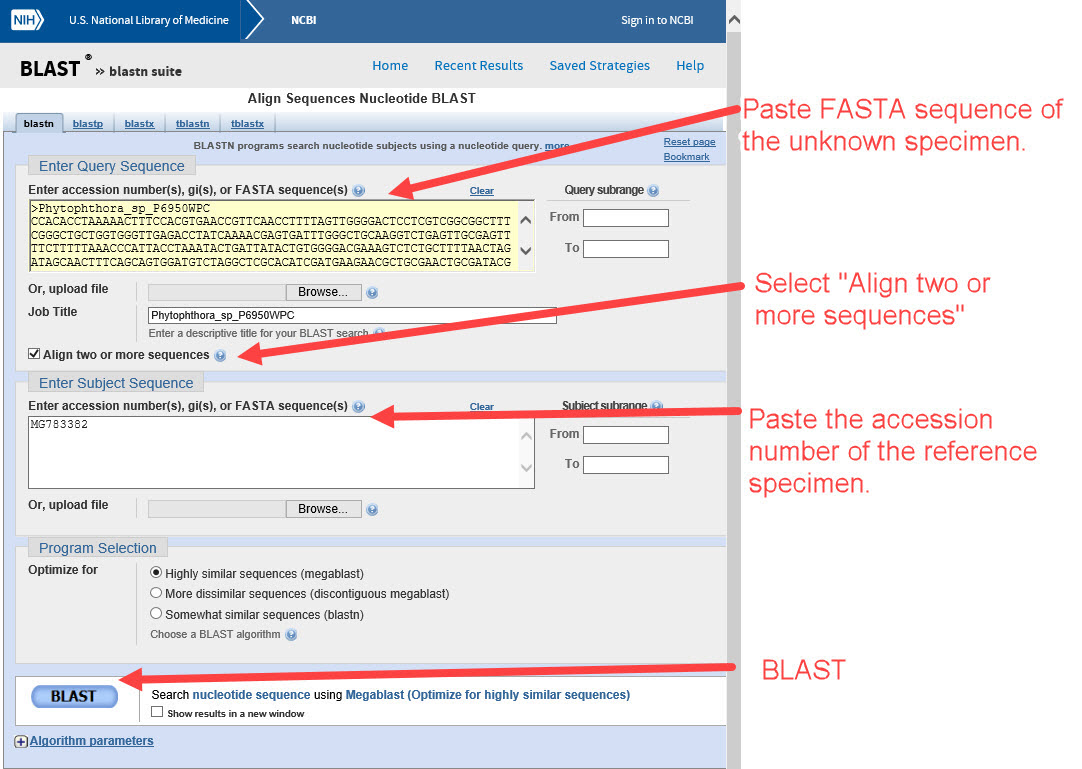
5) Go to NCBI BLAST blastn suite and select "Align two or more sequences". Paste the FASTA sequence of the Phytophthora unknown in the UPPER box and the accession number of the possible species match in the LOWER box (example P. boehmeriae in Clade 10, MG783382). Perform the BLAST and .... -
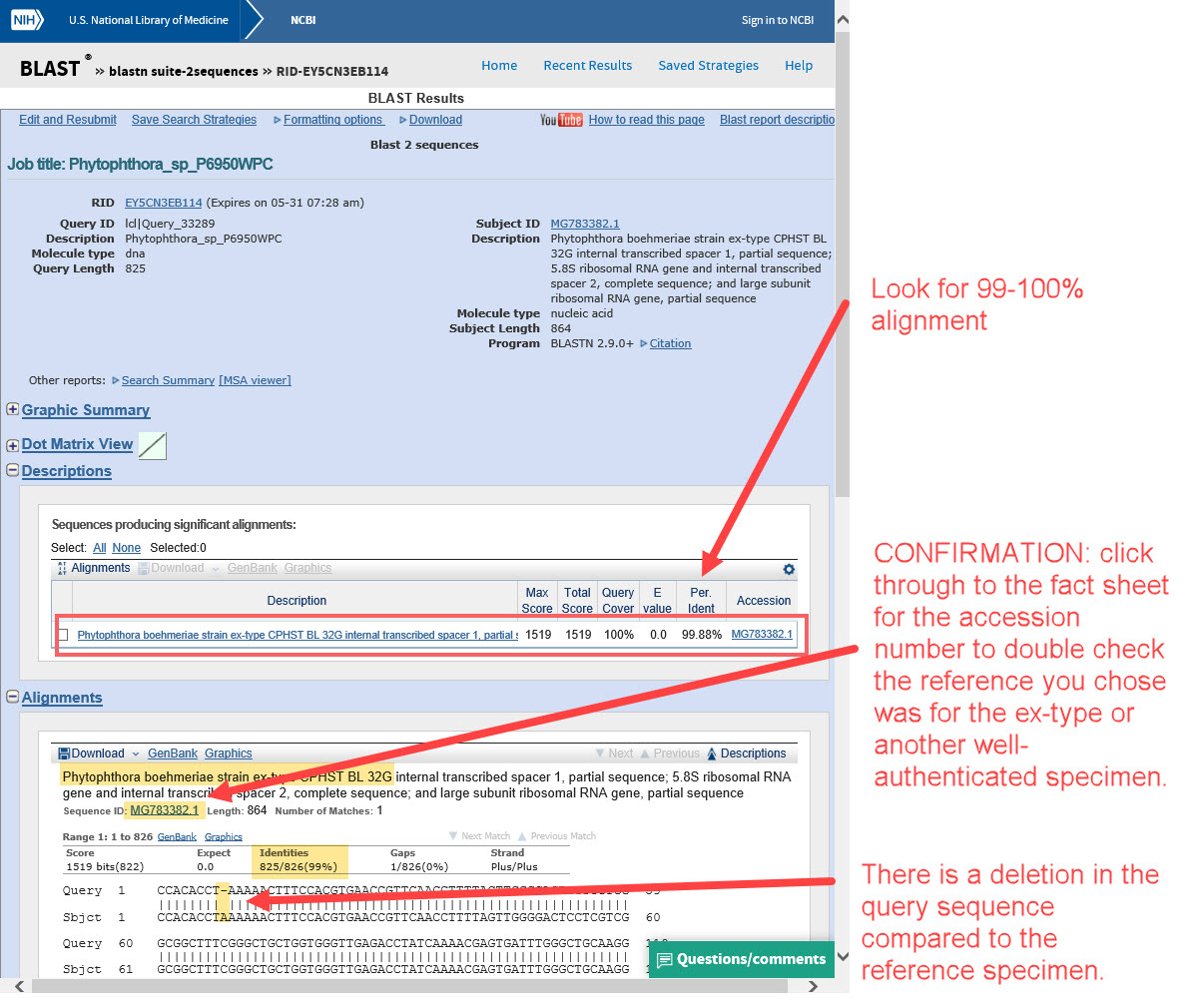
6) ... check the alignment. Does it align 99-100%? If so, we should confirm it’s a good reference specimen.
-
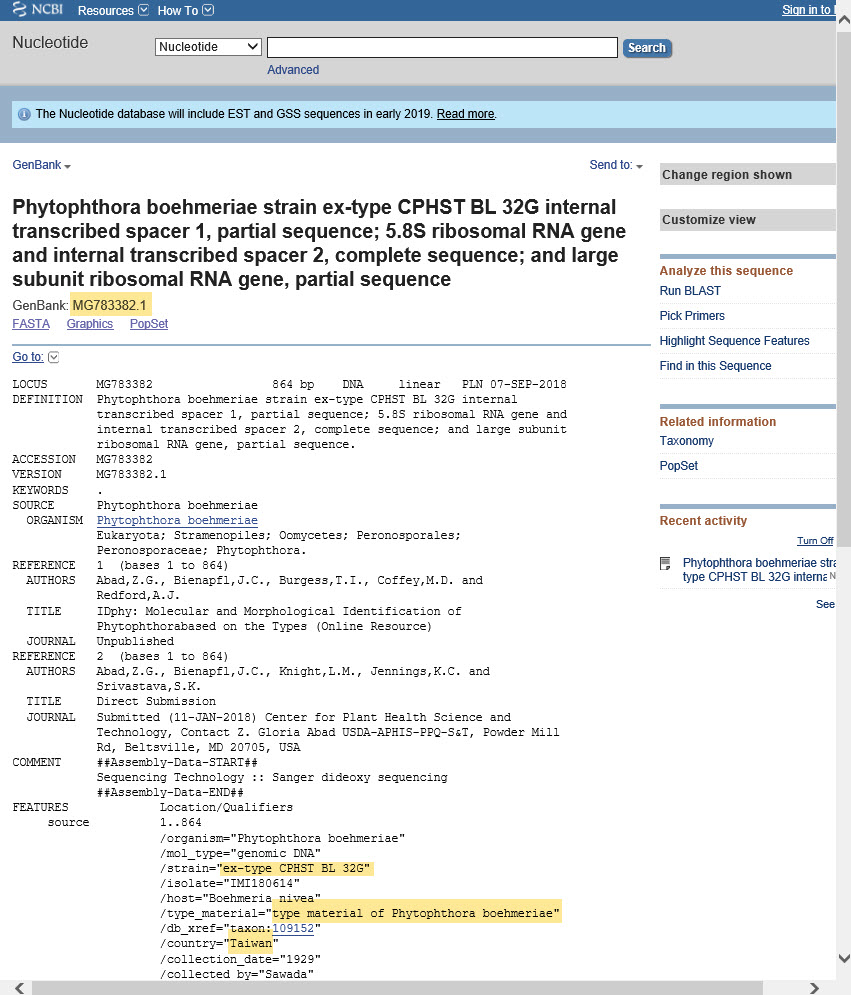
7) It is very important to verify that the reference sequence is from a type or other well-authenticated specimen. Check the fact sheet for the accession number you used as the reference for your unknown (click the accession number link in the alignment results, as shown above).
Confirm identity using a full clade analysis
The example shown below uses sequence number 2 from SOP-PID-05.
-
8) Go to NCBI BLAST blastn suite "Align two or more sequences" and paste the FASTA sequence of the Phytophthora unknown in UPPER box. Copy and paste all accession numbers in the clade (from the appropriate accession number table) in the LOWER box (for this example, all species in clade 10). -
9) Check the alignment of the unknown/query with all species in the clade. Do you observe 99-100% alignment with the same species as before? Check the alignment for each of the closest species. The best match appears to be P. gondwanensis, but before we select the best match, we should evaluate using a draft distance tree. -
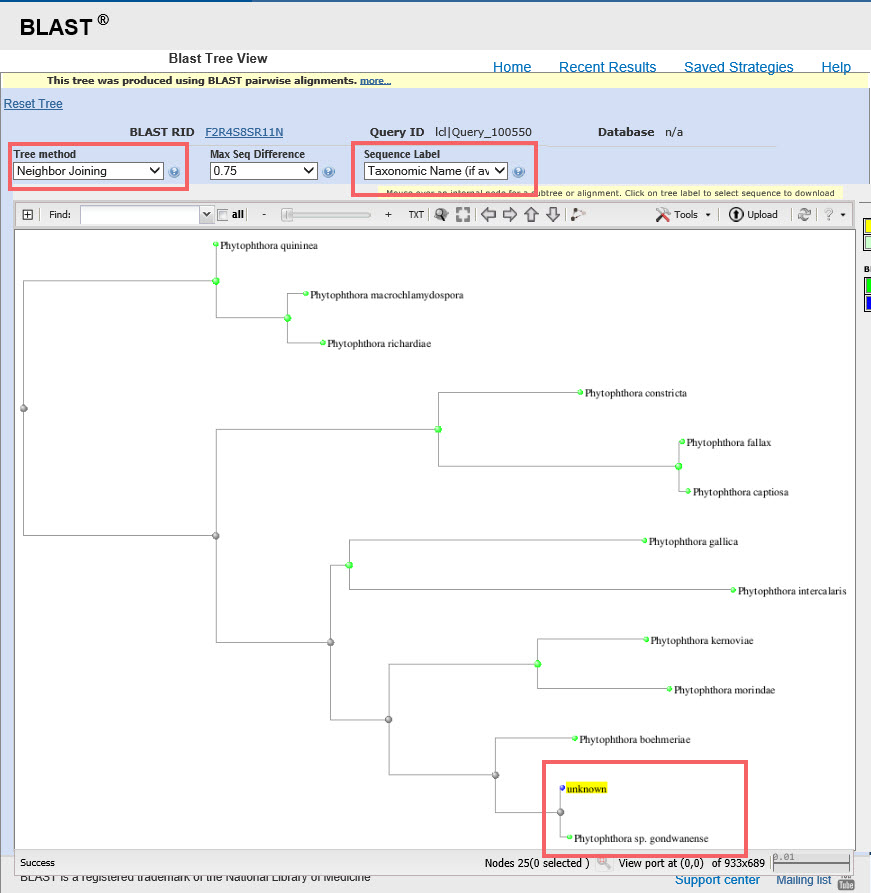
10) Prepare a distance tree through NCBI. In the results for the full clade multi-sequence alignment, click “Distance tree of results” under “Other reports.” For “Tree method,” select “Neighbor joining.” For “Sequence label,” select “Taxonomic name.” Unknown will be marked in yellow. This example shows the best match is P. gondwanensis, as confirmed by the distance tree.
Conclusion
If you follow steps 1-10 with an unknown specimen, and each test matches the unknown with the same result, and that match is from a type or another well-authenticated specimen, then you can confirm that the ITS rDNA sequence for your specimen is a particular species, based on comparison with sequences of the ex-type and other types in the same clade.
Accession number tables
-
ITS and COI accession numbers for Phytophthora species in clade 1 -
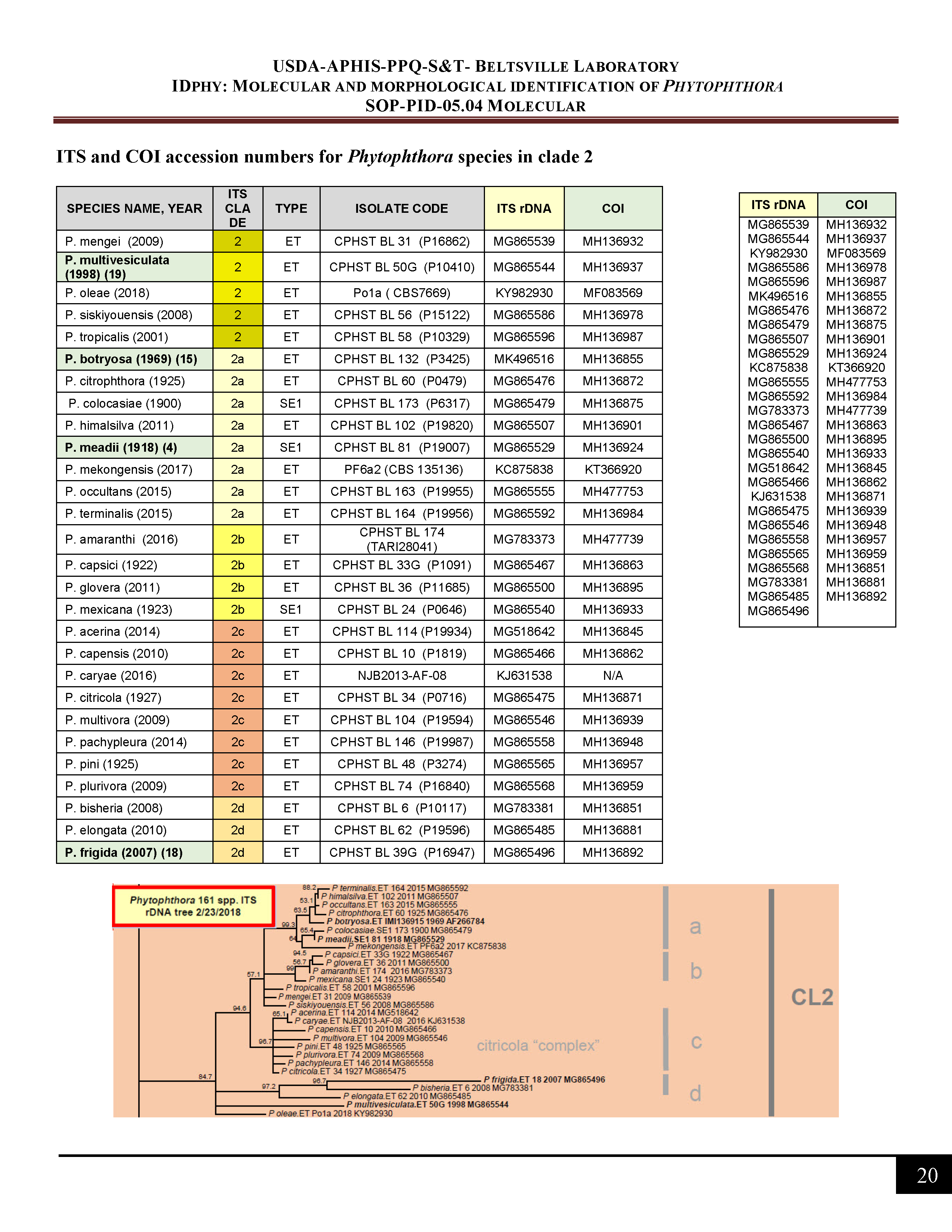
ITS and COI accession numbers for Phytophthora species in clade 2 -
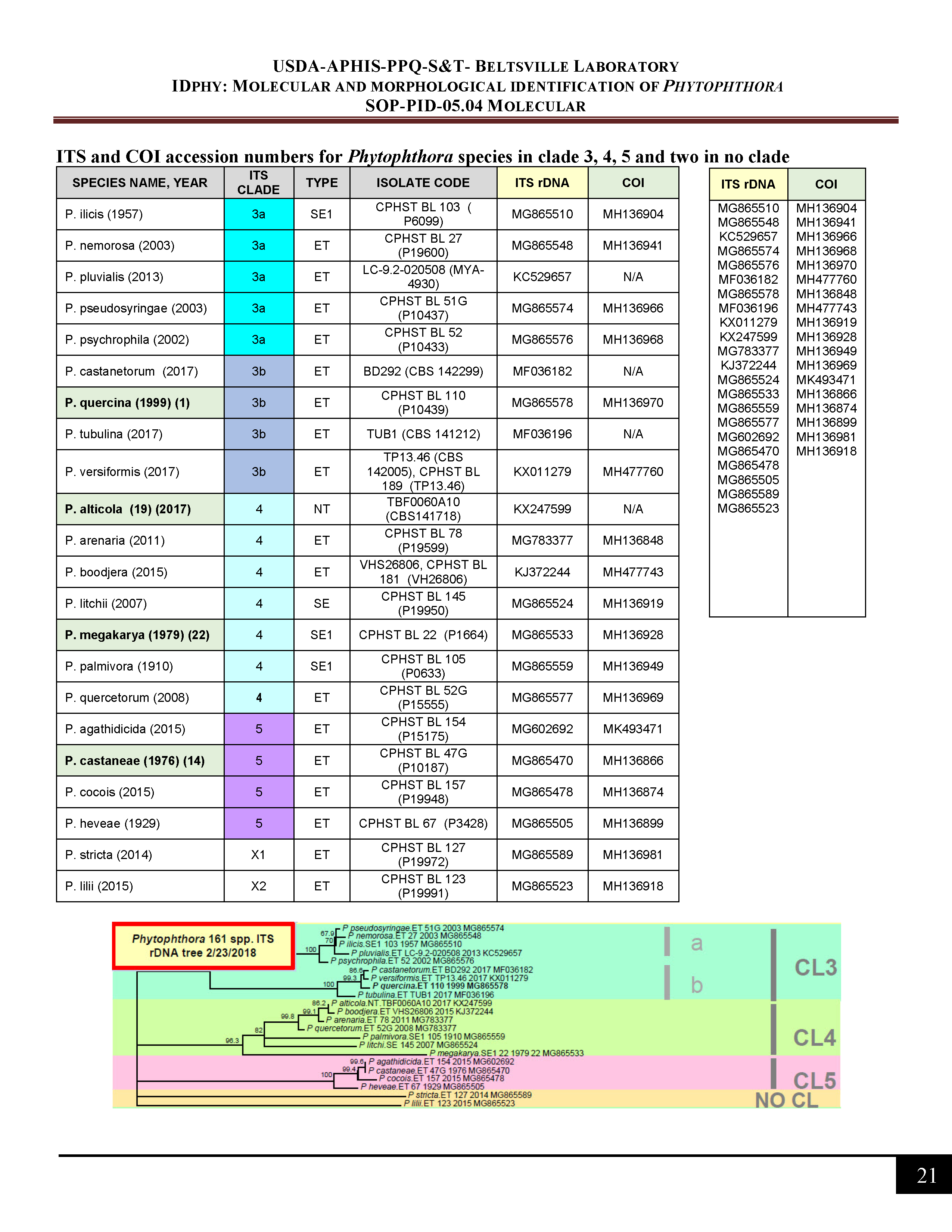
ITS and COI accession numbers for Phytophthora species in clade 3, 4, 5 and no clade -
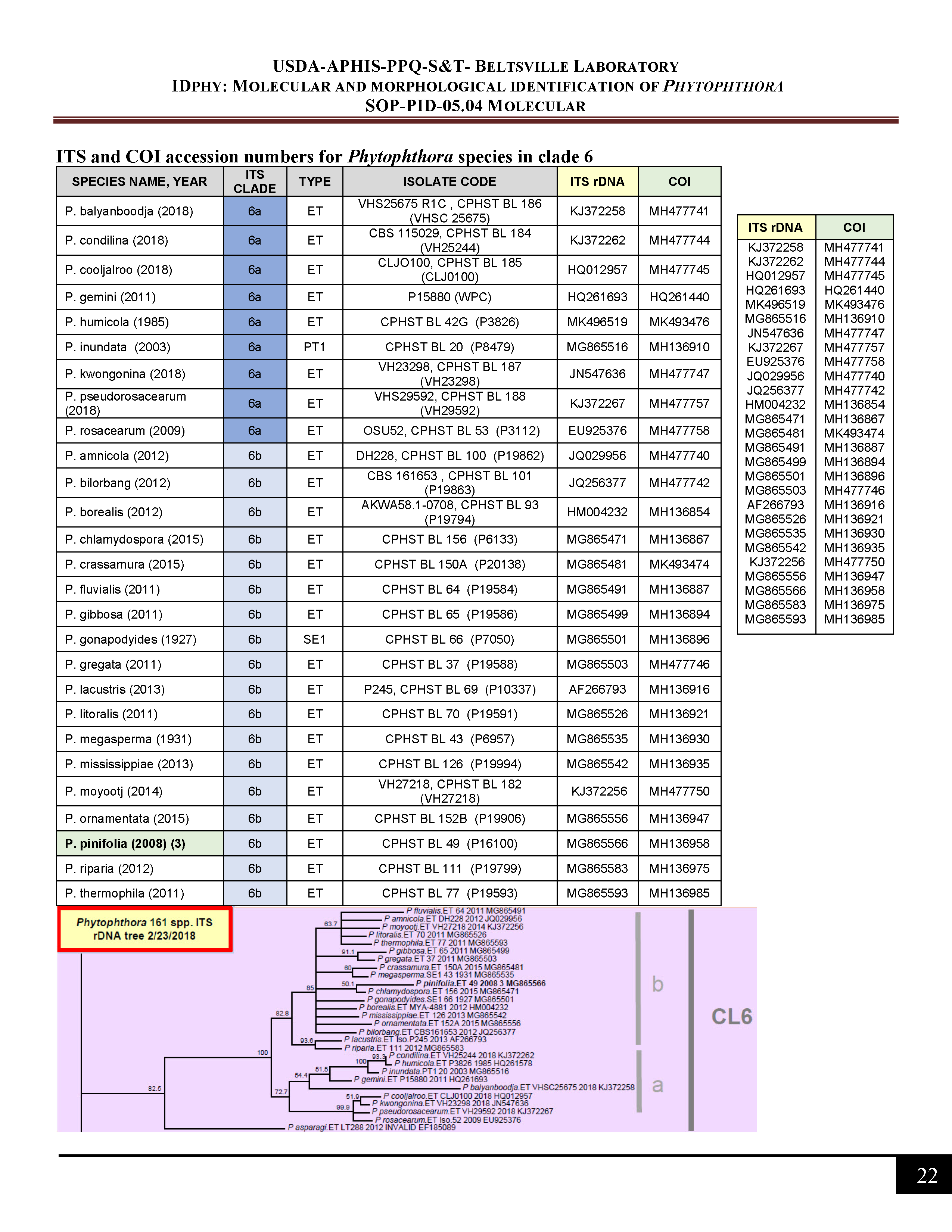
ITS and COI accession numbers for Phytophthora species in clade 6 -
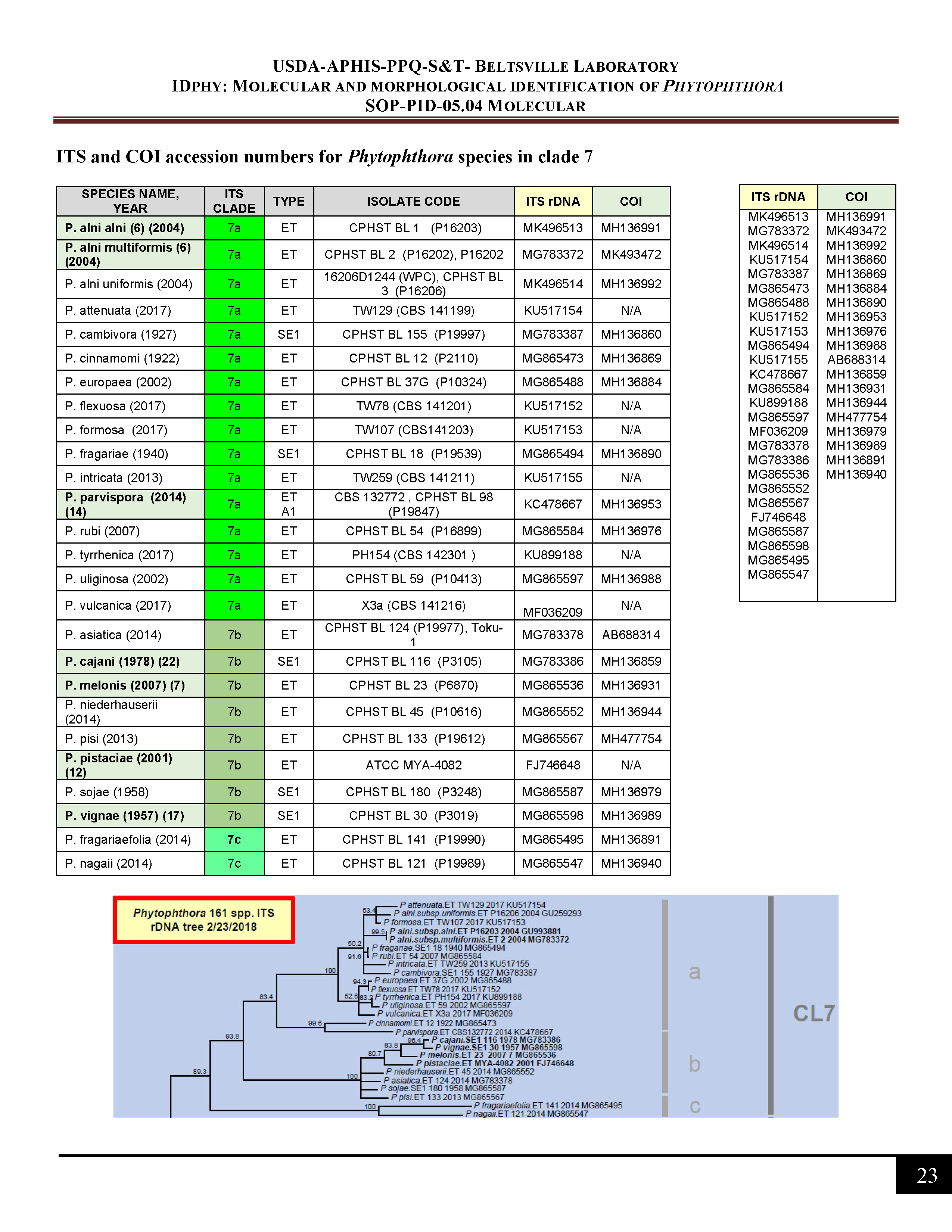
ITS and COI accession numbers for Phytophthora species in clade 7 -
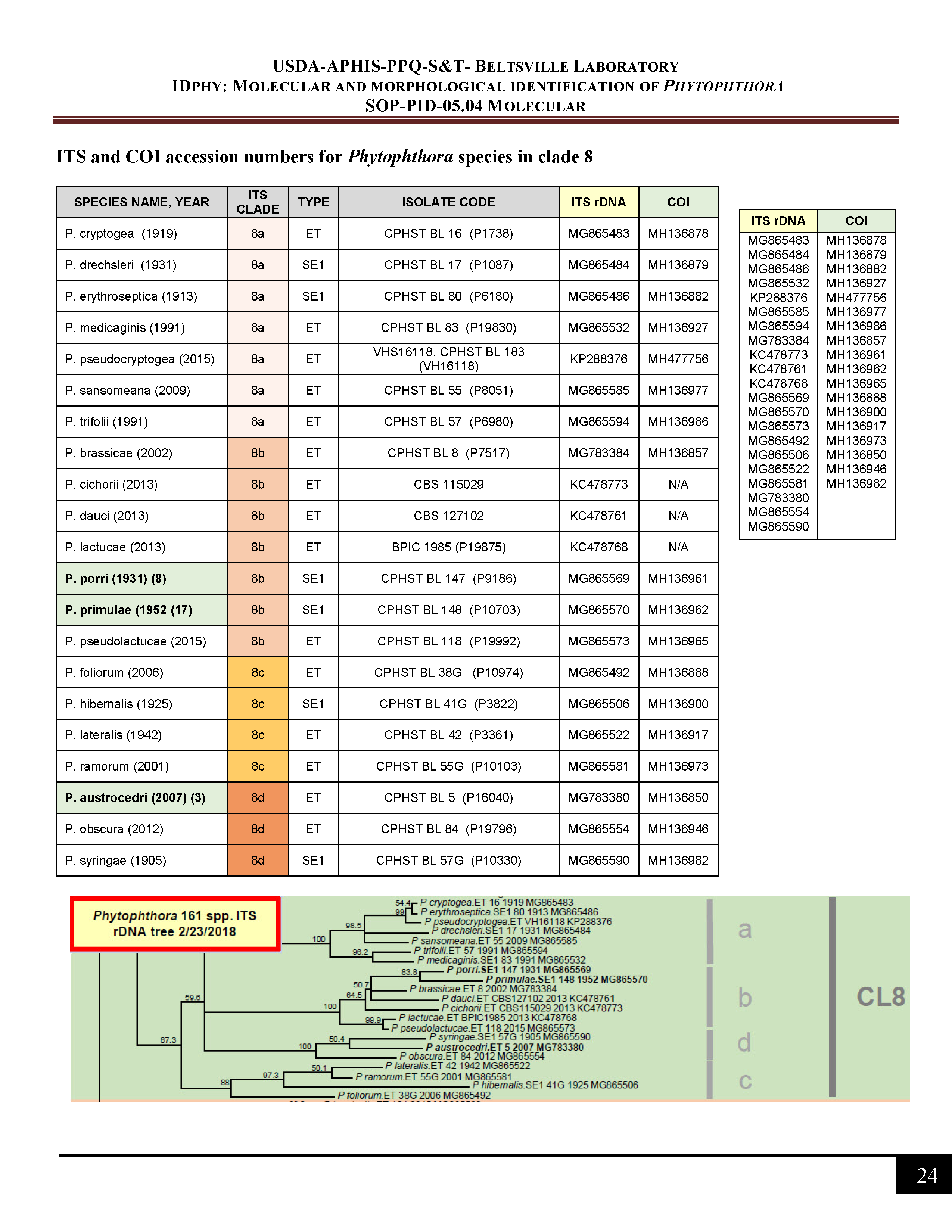
ITS and COI accession numbers for Phytophthora species in clade 8 -
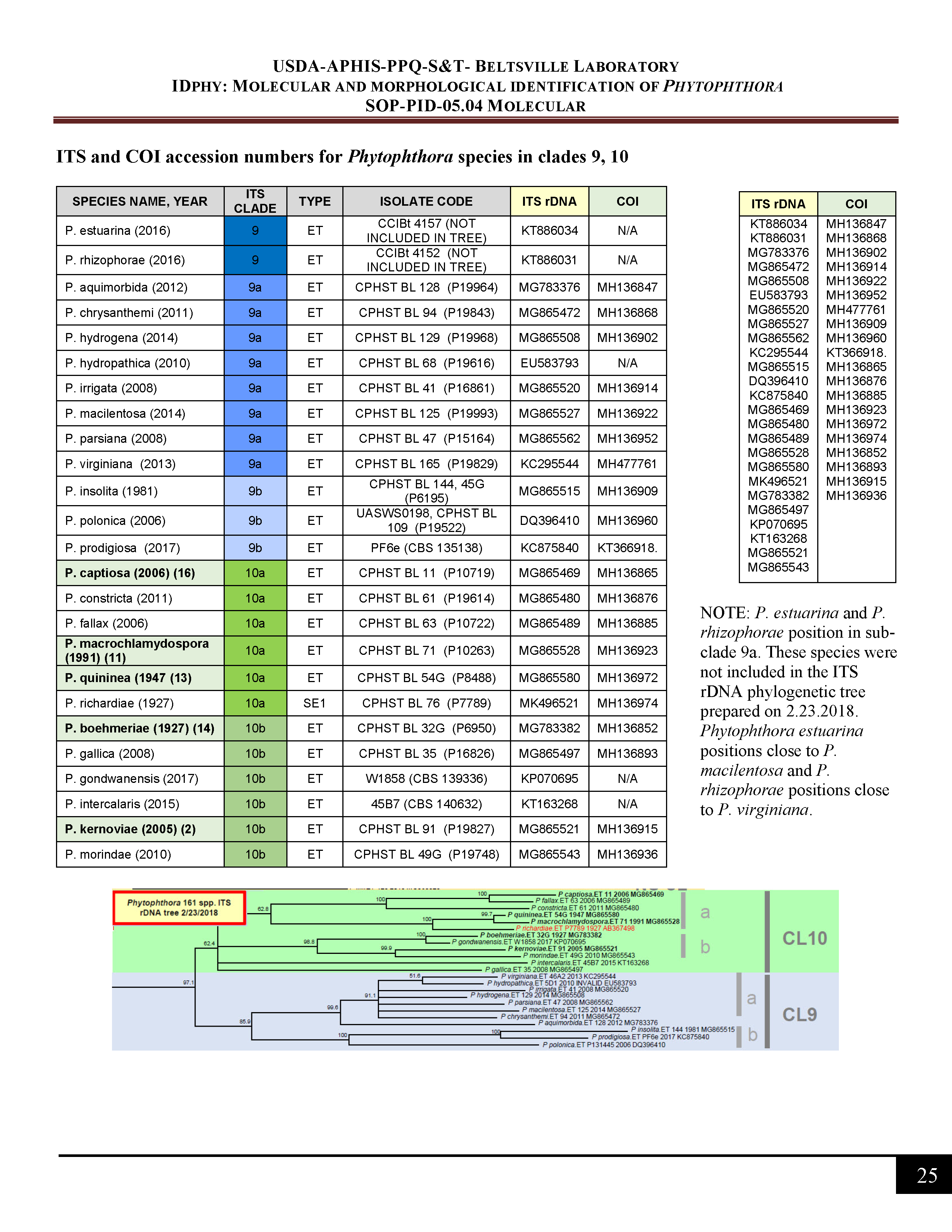
ITS and COI accession numbers for Phytophthora species in clade 9, 10